Structure, dynamics, and other biophysical properties of proteins, nanoparticles, and polymers interacting with membranes are investigated using all-atom and coarse-grained molecular dynamics (MD) simulations. The work aims to help the rational design and control of cell-targeting nanoparticles of protein drugs, self-assembled fibrous biomaterials and synthetic biology, and nanopore-based biosensors.
1. PEGylated particles interacting with lipid membranes
1.1 Dendrimer : conformation, internal structure, and cytotoxicity
1.2 Single-walled carbon nanotube : cytotoxicity and mushroom-brush transition
2. Biomolecules interacting with polymers and lipid membranes
2.1 Antimicrobial peptides: aggregation & toxicity 2.2 Assembly of PEGylated coiled-coil peptides
2.3 Others (siRNA, human serum albumin, lung-surfactant peptide, thermosensitive polypeptide)
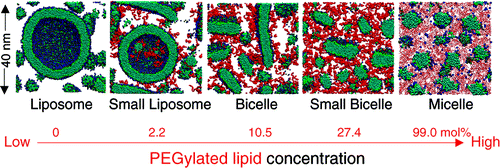
3. Self-assembly of liposomes, bicelles, and micelles: effect of PEGylation on the phase behavior
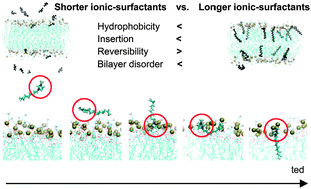
4. Imidazolium-based ionic liquids interacting with gramicidin A and lipid membranes
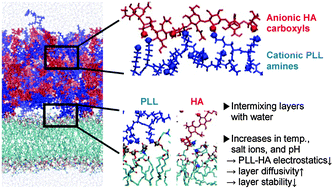
5. Layer-by-layer films of hyaluronic acids and poly-L-lysines: effects of pH, salt, and molecular weight
6. Free energy calculation 7. Force-field parameterization